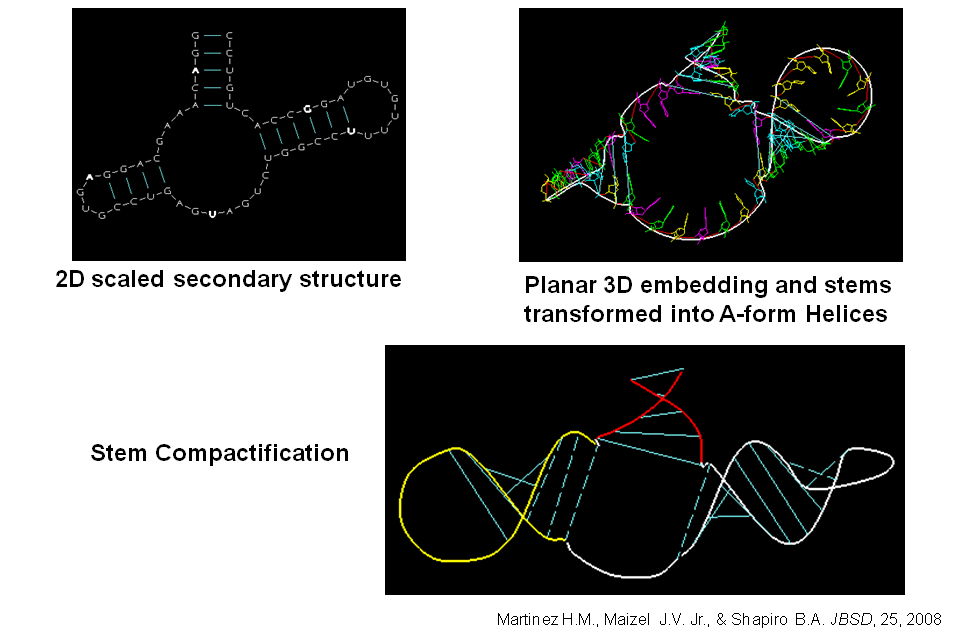
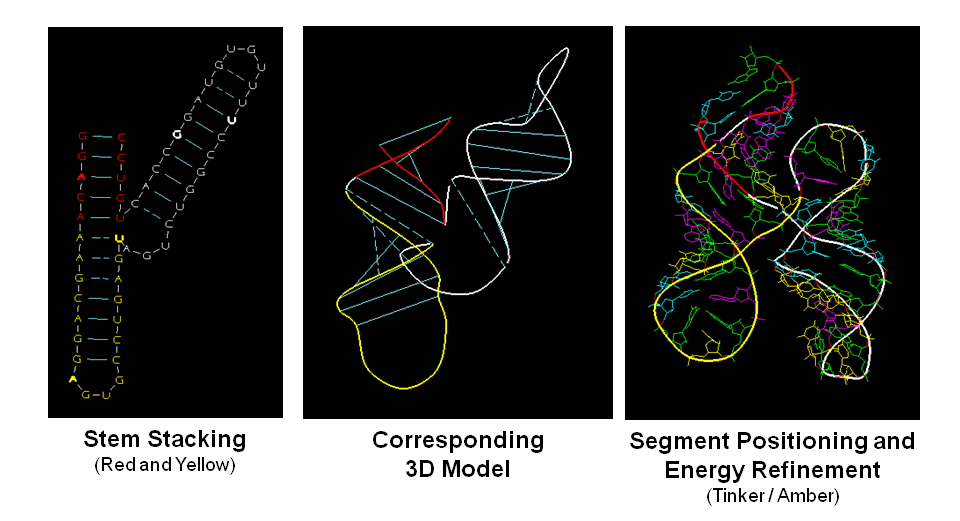
The program RNA2D3D accepts the primary RNA sequence information and a secondary structure representation for the sequence, which may include pseudoknots (Martinez et al. 2008). It rapidly generates a first-pass, three-dimensional model from the secondary structure. Further refinements of the models are usually necessary, but in many cases the first pass model will give significant insights as to how to proceed. Because of its speed and because of the flexibility of its structure editing tools, RNA2D3D can be used to quickly explore alternative 3D conformations in an interactive fashion (Note that it is not a de novo 3D structure prediction program.). For an example, see the tRNA-Shaped Structure in TCV 3' UTR.
The following list of steps captures the intended use approach:
- Start with RNA secondary and possible tertiary interaction data.
- Apply RNA2D3D to this data to produce a rough approximation of the 3D model.
- Check for steric clashes and adjust using editing tools if necessary.
- Apply the molecular mechanics minimization to spot any missed steric clashes and to improve bonded and non-bonded interactions.
- Check for conservation of given secondary and tertiary structure interactions and adjust using editing tools if necessary.
- For complex structures, check PDB database for similar structures. If necessary, adjust the model manually and/or splice in a found database motif.
- If additional tertiary structure interactions are found, manually adjust and go to step 4. Otherwise, go to step 8.
- Apply molecular dynamics. If you are satisfied, STOP. If not, go to step 7.
DOWNLOAD SOFTWARE HERE.