Molecular Modeling Study of Human Telomerase RNA and implications of mutations
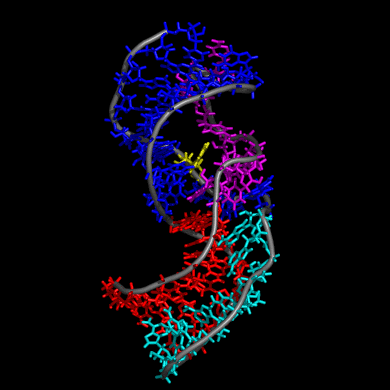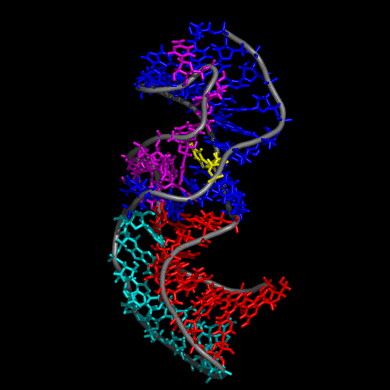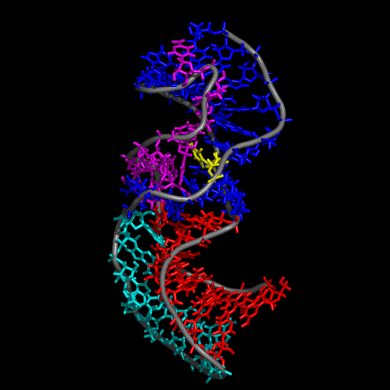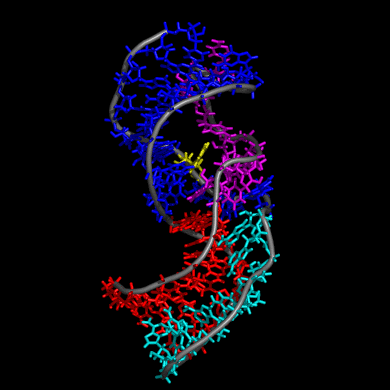
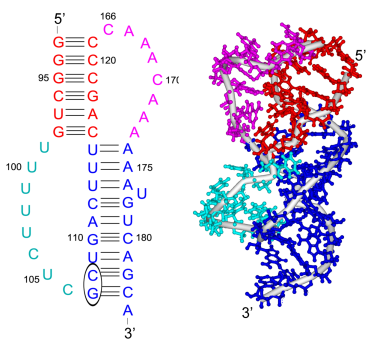
Our study shows that the wild-type telomerase RNA hairpin is very flexible with periodic exhibition of dramatic structural fluctuations represented by the opening and closing of a non-canonical base pair region. These structural deviations correspond to significant disruptions of the direct hydrogen bonding network in the helix, widening of the major groove of the hairpin structure, and causing several U and C nucleotides to protrude into the major groove from the helix. We suggest that these structural fluctuations expose a nucleation point for pseudoknot formation. The DKC mutations stabilize the hairpin dynamics due to base stacking in the pentaloop.
We also created a simulated telomerase pseudoknot structure and examined its folding and formation of various tertiary interactions. The DKC mutations of the 3D pseudoknot structure revealed significant change in stability and disruption of hydrogen bonding in the pseudoknot P3 region.